réservé à la recherche
I-BET151 (GSK1210151A) BET inhibiteur
N° Cat.S2780
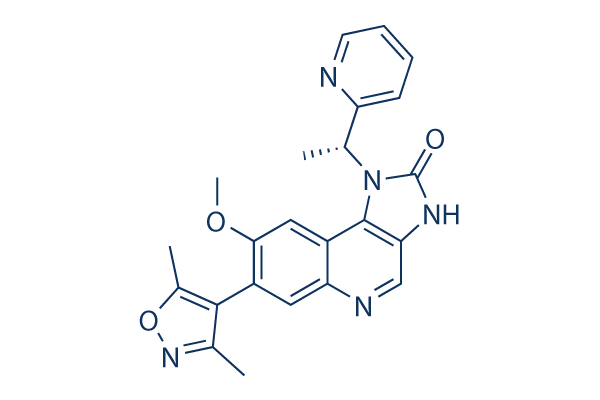
Structure chimique
Poids moléculaire: 415.44
Contrôle qualité
Culture cellulaire, traitement et concentration de travail
| Lignées cellulaires | Type dessai | Concentration | Temps dincubation | Formulation | Description de lactivité | PMID |
|---|---|---|---|---|---|---|
| MV4;11 | cytotoxicity assay | ~100 μM | DMSO | IC50=26 nM | 21964340 | |
| RS4;11 | cytotoxicity assay | ~100 μM | DMSO | IC50=192 nM | 21964340 | |
| MOLM13 | cytotoxicity assay | ~100 μM | DMSO | IC50=120 nM | 21964340 | |
| NOMO1 | cytotoxicity assay | ~100 μM | DMSO | IC50=15 nM | 21964340 | |
| HEL | cytotoxicity assay | ~100 μM | DMSO | IC50=1 μM | 21964340 | |
| K562 | cytotoxicity assay | ~100 μM | DMSO | IC50>100 μM | 21964340 | |
| MEG01 | cytotoxicity assay | ~100 μM | DMSO | IC50=25 μM | 21964340 | |
| HL60 | cytotoxicity assay | ~100 μM | DMSO | IC50=890 nM | 21964340 | |
| MV4;11 | Apoptosis assay | ~100 μM | DMSO | induces apoptosis | 21964340 | |
| MOLM13 | Apoptosis assay | ~100 μM | DMSO | induces apoptosis | 21964340 | |
| MV4;11 | Function assay | DMSO | decreases the recruitment of BRD3/4 and impaired recruitment of CDK9 and PAF1 to the transcriptional start site | 21964340 | ||
| PBMC | Function assay | DMSO | inhibits IL-6 with pIC50 of 6.7 | 22437115 | ||
| A2 | Function assay | ~10 μM | DMSO | reactivates latent HIV-1 | 23255218 | |
| A72 | Function assay | ~10 μM | DMSO | reactivates latent HIV-1 | 23255218 | |
| BC1 | Growth inhibitory assay | ~1 μM | DMSO | IC50=220 nM | 23792448 | |
| BC3 | Growth inhibitory assay | ~1 μM | DMSO | IC50=460 nM | 23792448 | |
| BCBL1 | Growth inhibitory assay | ~1 μM | DMSO | IC50=330 nM | 23792448 | |
| BJAB | Growth inhibitory assay | ~1 μM | DMSO | IC50=970 nM | 23792448 | |
| Namalwa | Growth inhibitory assay | ~1 μM | DMSO | IC50=970 nM | 23792448 | |
| Jurkat | Growth inhibitory assay | ~1 μM | DMSO | IC50=1220 nM | 23792448 | |
| MM1S | Growth inhibitory assay | ~1 μM | DMSO | IC50=760 nM | 23792448 | |
| U266 | Growth inhibitory assay | ~1 μM | DMSO | IC50=950 nM | 23792448 | |
| UM-PEL-1 | Growth inhibitory assay | ~1 μM | DMSO | IC50=210 nM | 23792448 | |
| UM-PEL-3 | Growth inhibitory assay | ~1 μM | DMSO | IC50=180 nM | 23792448 | |
| BC1 | Function assay | 500 nM | DMSO | induces cell-cycle arrest | 23792448 | |
| BC3 | Function assay | 500 nM | DMSO | induces cell-cycle arrest | 23792448 | |
| BC1 | Function assay | 800 nM | DMSO | reduces c-Myc protein levels | 23792448 | |
| BC3 | Function assay | 800 nM | DMSO | reduces c-Myc protein levels | 23792448 | |
| H929 | Function assay | ~1 μM | DMSO | induces cell cycle arrest | 24335499 | |
| KMS12PE | Function assay | ~1 μM | DMSO | induces cell cycle arrest | 24335499 | |
| KMS12BM | Function assay | ~1 μM | DMSO | induces cell cycle arrest | 24335499 | |
| KMS18 | Function assay | ~1 μM | DMSO | induces cell cycle arrest | 24335499 | |
| KMS11 | Function assay | ~1 μM | DMSO | induces cell cycle arrest | 24335499 | |
| RPMI8226 | Function assay | ~1 μM | DMSO | induces cell cycle arrest | 24335499 | |
| H929 | Apoptosis assay | ~1 μM | DMSO | induces cell apoptosis | 24335499 | |
| KMS12PE | Apoptosis assay | ~1 μM | DMSO | induces cell apoptosis | 24335499 | |
| KMS12BM | Apoptosis assay | ~1 μM | DMSO | induces cell apoptosis | 24335499 | |
| KMS18 | Apoptosis assay | ~1 μM | DMSO | induces cell apoptosis | 24335499 | |
| KMS11 | Apoptosis assay | ~1 μM | DMSO | induces cell apoptosis | 24335499 | |
| RPMI8226 | Apoptosis assay | ~1 μM | DMSO | induces cell apoptosis | 24335499 | |
| U87MG | Function assay | ~10 μM | DMSO | reduces U87MG cellular ATP with IC50 of 1.05 μM | 24496381 | |
| A172 | Function assay | ~10 μM | DMSO | reduces cellular ATP with IC50 of 1.28 μM | 24496381 | |
| SW1783 | Function assay | ~10 μM | DMSO | reduces cellular ATP with IC50 of 2.68 μM | 24496381 | |
| U87MG | Function assay | ~10 μM | DMSO | increases proportion of cells in the G1/S transition | 24496381 | |
| RAW267.4 | Function assay | 1 μM | DMSO | reduces IL-6 production induced by LPS | 24859008 | |
| RAW267.4 | Function assay | 1 μM | DMSO | reduces the association between BRD4 and acetylated p65 | 24859008 | |
| Me007 | Growth inhibitory assay | ~100 μM | DMSO | inhibits the growth | 24906137 | |
| SK-Mel-28 | Growth inhibitory assay | ~100 μM | DMSO | inhibits the growth | 24906137 | |
| Mel-RMU | Growth inhibitory assay | ~100 μM | DMSO | inhibits the growth | 24906137 | |
| Mel-JD | Growth inhibitory assay | ~100 μM | DMSO | inhibits the growth | 24906137 | |
| Mel-RM | Growth inhibitory assay | ~100 μM | DMSO | inhibits the growth | 24906137 | |
| Me007 | Apoptosis assay | ~100 μM | DMSO | induces apoptosis | 24906137 | |
| SK-Mel-28 | Apoptosis assay | ~100 μM | DMSO | induces apoptosis | 24906137 | |
| Mel-RMU | Apoptosis assay | ~100 μM | DMSO | induces apoptosis | 24906137 | |
| Mel-JD | Apoptosis assay | ~100 μM | DMSO | induces apoptosis | 24906137 | |
| Mel-RM | Apoptosis assay | ~100 μM | DMSO | induces apoptosis | 24906137 | |
| Me007 | Function assay | 10 μM | DMSO | induces cell cycle arrest by upregulation of p21 | 24906137 | |
| SK-Mel-28 | Function assay | 10 μM | DMSO | induces cell cycle arrest by upregulation of p21 | 24906137 | |
| Mel-RMU | Function assay | 10 μM | DMSO | induces cell cycle arrest by upregulation of p21 | 24906137 | |
| Mel-JD | Function assay | 10 μM | DMSO | induces cell cycle arrest by upregulation of p21 | 24906137 | |
| Mel-RM | Function assay | 10 μM | DMSO | induces cell cycle arrest by upregulation of p21 | 24906137 | |
| Me007 | Function assay | 10 μM | DMSO | upregulates proapoptotic and cell cycle arrest genes | 24906137 | |
| SK-Mel-28 | Function assay | 10 μM | DMSO | upregulates proapoptotic and cell cycle arrest genes | 24906137 | |
| Mel-RMU | Function assay | 10 μM | DMSO | upregulates proapoptotic and cell cycle arrest genes | 24906137 | |
| Mel-JD | Function assay | 10 μM | DMSO | upregulates proapoptotic and cell cycle arrest genes | 24906137 | |
| Mel-RM | Function assay | 10 μM | DMSO | upregulates proapoptotic and cell cycle arrest genes | 24906137 | |
| HepG2 | Function assay | 18 hrs | Upregulation of ApoA1 expression in human HepG2 cells assessed as concentration required to increase 70% of luciferase activity after 18 hrs by luciferase reporter gene assay, EC170 = 0.09 μM. | 22386529 | ||
| Raji | Function assay | 4 hrs | Inhibition of BRD4 in human Raji cells assessed as reduction of MYC expression after 4 hrs, IC50 = 0.13 μM. | 24900758 | ||
| MV4-11 | Growth inhibition assay | 72 hrs | Growth inhibition of human MV4-11 cells after 72 hrs by SRB assay, IC50 = 0.119 μM. | 25559428 | ||
| MM1S | Growth inhibition assay | 72 hrs | Growth inhibition of human MM1S cells after 72 hrs by SRB assay, IC50 = 0.299 μM. | 25559428 | ||
| HT-29 | Growth inhibition assay | 72 hrs | Growth inhibition of human HT-29 cells after 72 hrs by SRB assay, IC50 = 0.945 μM. | 25559428 | ||
| Rosetta2 DE3 | Function assay | 30 mins | Displacement of FAM-labeled ZBA248 from BRD4 BD1 (44 to 168 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by fluorescence polarization assay, IC50 = 0.0317 μM. | 26080064 | ||
| MV4-11 | Cytotoxicity assay | 4 days | Cytotoxicity against human MV4-11 cells harboring MLL1 fusion gene assessed as growth inhibition after 4 days by CellTiter-Glo luminescent assay, IC50 = 0.162 μM. | 26080064 | ||
| Rosetta2 DE3 | Function assay | 30 mins | Displacement of FAM-labeled ZBA248 from BRD4 BD2 (333 to 460 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by fluorescence polarization assay, IC50 = 0.226 μM. | 26080064 | ||
| MOLM13 | Cytotoxicity assay | 4 days | Cytotoxicity against human MOLM13 cells harboring MLL1 fusion gene assessed as growth inhibition after 4 days by CellTiter-Glo luminescent assay, IC50 = 0.228 μM. | 26080064 | ||
| Rosetta2 DE3 | Function assay | 30 mins | Inhibition of FAM-labeled ZBA248 binding to recombinant human N-terminal His6-tagged BRD4 bromodomain 1 (44 to 168 residues) expressed in Rosetta2 DE3 cells after 30 mins by Flourescence polarization assay, IC50 = 0.0317 μM. | 28463487 | ||
| MV4-11 | Growth inhibition assay | 4 days | Growth inhibition of human MV4-11 cells after 4 days by WST-8 assay, IC50 = 0.162 μM. | 28463487 | ||
| Rosetta2 DE3 | Function assay | 30 mins | Inhibition of FAM-labeled ZBA248 binding to recombinant human N-terminal His6-tagged BRD4 bromodomain 2 (333 to 460 residues) expressed in Rosetta2 DE3 cells after 30 mins by Flourescence polarization assay, IC50 = 0.226 μM. | 28463487 | ||
| MOLM13 | Growth inhibition assay | 4 days | Growth inhibition of human MOLM13 cells after 4 days by WST-8 assay, IC50 = 0.228 μM. | 28463487 | ||
| Rosetta2 DE3 | Function assay | Binding affinity to biotinylated BRD3 BD1 (24 to 144 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells by bio-layer interferometry method, Kd = 0.0298 μM. | 26080064 | |||
| Rosetta2 DE3 | Function assay | Binding affinity to biotinylated BRD3 BD2 (306 to 417 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells by bio-layer interferometry method, Kd = 0.0405 μM. | 26080064 | |||
| Rosetta2 DE3 | Function assay | Binding affinity to biotinylated BRD4 BD1 (44 to 168 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells by bio-layer interferometry method, Kd = 0.0528 μM. | 26080064 | |||
| Rosetta2 DE3 | Function assay | Binding affinity to biotinylated BRD2 BD1 (72 to 205 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells by bio-layer interferometry method, Kd = 0.0548 μM. | 26080064 | |||
| Rosetta2 DE3 | Function assay | Binding affinity to biotinylated BRD2 BD2 (349 to 460 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells by bio-layer interferometry method, Kd = 0.0703 μM. | 26080064 | |||
| Rosetta2 DE3 | Function assay | Binding affinity to biotinylated BRD4 BD2 (333 to 460 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells by bio-layer interferometry method, Kd = 0.215 μM. | 26080064 | |||
| Rosetta2 DE3 | Function assay | Binding affinity to biotinylated CREBBP (1043 to 1159 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells by bio-layer interferometry method, Kd = 3.084 μM. | 26080064 | |||
| Rosetta2 DE3 | Function assay | 30 mins | Displacement of FAM-labeled ZBA248 from BRD3 BD1 (24 to 144 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by fluorescence polarization assay, Ki = 0.0072 μM. | 26080064 | ||
| Rosetta2 DE3 | Function assay | 30 mins | Displacement of FAM-labeled ZBA248 from BRD4 BD1 (44 to 168 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by fluorescence polarization assay, Ki = 0.009 μM. | 26080064 | ||
| Rosetta2 DE3 | Function assay | 30 mins | Displacement of FAM-labeled ZBA248 from BRD2 BD1 (72 to 205 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by fluorescence polarization assay, Ki = 0.009 μM. | 26080064 | ||
| Rosetta2 DE3 | Function assay | 30 mins | Displacement of FAM-labeled ZBA248 from BRD3 BD2 (306 to 417 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by fluorescence polarization assay, Ki = 0.0223 μM. | 26080064 | ||
| Rosetta2 DE3 | Function assay | 30 mins | Displacement of FAM-labeled ZBA248 from BRD2 BD2 (349 to 460 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by fluorescence polarization assay, Ki = 0.0496 μM. | 26080064 | ||
| Rosetta2 DE3 | Function assay | 30 mins | Displacement of FAM-labeled ZBA248 from BRD4 BD2 (333 to 460 amino acid residues) (unknown origin) expressed in Rosetta2 DE3 cells after 30 mins by fluorescence polarization assay, Ki = 0.0748 μM. | 26080064 | ||
| Rosetta2 DE3 | Function assay | 30 mins | Inhibition of FAM-labeled ZBA248 binding to recombinant human N-terminal His6-tagged BRD4 bromodomain 1 (44 to 168 residues) expressed in Rosetta2 DE3 cells after 30 mins by Flourescence polarization assay, Ki = 0.009 μM. | 28463487 | ||
| Rosetta2 DE3 | Function assay | 30 mins | Inhibition of FAM-labeled ZBA248 binding to recombinant human N-terminal His6-tagged BRD4 bromodomain 2 (333 to 460 residues) expressed in Rosetta2 DE3 cells after 30 mins by Flourescence polarization assay, Ki = 0.0748 μM. | 28463487 | ||
| THP1 | Antiinflammatory assay | Antiinflammatory activity in human THP1 cells | 22386529 | |||
| HT-29 | Function assay | 0.3125 uM to 5 uM | 24 hrs | Inhibition of BRD4 in human HT-29 cells assessed as reduction in c-Myc protein expression at 0.3125 uM to 5 uM uM after 24 hrs by Western blotting method | 25559428 | |
| MV4-11 | Function assay | Inhibition of BRD4 in human MV4-11 cells assessed as downregulation of BCL2 RNA expression by RNA-seq analysis | 29259751 | |||
| MV4-11 | Function assay | Inhibition of BRD4 in human MV4-11 cells assessed as downregulation of cMYC RNA expression by RNA-seq analysis | 29259751 | |||
| U-2 OS | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for U-2 OS cells | 29435139 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for A673 cells | 29435139 | |||
| DAOY | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for DAOY cells | 29435139 | |||
| BT-37 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for BT-37 cells | 29435139 | |||
| RD | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for RD cells | 29435139 | |||
| SK-N-SH | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-SH cells | 29435139 | |||
| BT-12 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for BT-12 cells | 29435139 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for MG 63 (6-TG R) cells | 29435139 | |||
| NB1643 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB1643 cells | 29435139 | |||
| OHS-50 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for OHS-50 cells | 29435139 | |||
| fibroblast cells | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for control Hh wild type fibroblast cells | 29435139 | |||
| Rh41 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Rh41 cells | 29435139 | |||
| Rh30 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Rh30 cells | 29435139 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SJ-GBM2 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-MC cells | 29435139 | |||
| NB-EBc1 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB-EBc1 cells | 29435139 | |||
| LAN-5 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for LAN-5 cells | 29435139 | |||
| Rh18 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Rh18 cells | 29435139 | |||
| Cliquez pour voir plus de données expérimentales sur les lignées cellulaires | ||||||
Informations chimiques, stockage et stabilité
| Poids moléculaire | 415.44 | Formule | C23H21N5O3 |
Stockage (À partir de la date de réception) | |
|---|---|---|---|---|---|
| N° CAS | 1300031-49-5 | Télécharger le SDF | Stockage des solutions mères |
|
|
| Synonymes | N/A | Smiles | CC1=C(C(=NO1)C)C2=C(C=C3C(=C2)N=CC4=C3N(C(=O)N4)C(C)C5=CC=CC=N5)OC | ||
Solubilité
|
In vitro |
DMSO
: 83 mg/mL
(199.78 mM)
Ethanol : 83 mg/mL Water : Insoluble |
Calculateur de molarité
|
In vivo |
|||||
Calculateur de formulation in vivo (Solution claire)
Étape 1 : Entrez les informations ci-dessous (Recommandé : Un animal supplémentaire pour tenir compte des pertes pendant lexpérience)
Étape 2 : Entrez la formulation in vivo (Ceci nest que le calculateur, pas la formulation. Veuillez nous contacter dabord sil ny a pas de formulation in vivo dans la section Solubilité.)
Résultats du calcul :
Concentration de travail : mg/ml;
Méthode de préparation du liquide maître DMSO : mg médicament prédissous dans μL DMSO ( Concentration du liquide maître mg/mL, Veuillez nous contacter dabord si la concentration dépasse la solubilité du DMSO du lot de médicament. )
Méthode de préparation de la formulation in vivo : Prendre μL DMSO liquide maître, ajouter ensuiteμL PEG300, mélanger et clarifier, ajouter ensuiteμL Tween 80, mélanger et clarifier, ajouter ensuite μL ddH2O, mélanger et clarifier.
Méthode de préparation de la formulation in vivo : Prendre μL DMSO liquide maître, ajouter ensuite μL Huile de maïs, mélanger et clarifier.
Remarque : 1. Assurez-vous que le liquide est clair avant dajouter le solvant suivant.
2. Assurez-vous dajouter le(s) solvant(s) dans lordre. Vous devez vous assurer que la solution obtenue lors de lajout précédent est une solution claire avant de procéder à lajout du solvant suivant. Des méthodes physiques telles que le vortex, les ultrasons ou le bain-marie peuvent être utilisées pour faciliter la dissolution.
Mécanisme daction
| Fonctionnalités |
Optimized to retain excellent BET target potency and selectivity while enhancing the in vivo pharmacokinetics and terminal half-life to enable prolonged in vivo studies.
|
|---|---|
| Targets/IC50/Ki |
BRD3
(Cell-free assay) 0.25 μM
BRD2
(Cell-free assay) 0.5 μM
BRD4
(Cell-free assay) 0.79 μM
|
| In vitro |
I-BET151 (GSK1210151A) présente une sélectivité puissante sur une vaste gamme de types de protéines diverses telles que COX-2, P450, Aurora B, GSK3β, PI3K-γ, GPCR, les canaux ioniques et les transporteurs. Similaire à I-BET762 (GSK525762A), il présente une forte affinité de liaison pour BRD2, BRD3 et BRD4 avec un KD de 0,02-0,1 μM, et inhibe significativement la production de cytokine IL-6 stimulée par le lipopolysaccharide dans les cellules mononucléaires du sang périphérique humain (PBMC) et le sang total (WB) ainsi que le WB de rat avec une IC50 de 0,16 μM, 1,26 μM et 1,26 μM, respectivement. Ce composé (0,5 ou 5 μM) inhibe la liaison des BET (BRD2, BRD3, BRD4 et BRD9) mais pas la liaison de 23 autres protéines à bromodomaine dans l'extrait nucléaire HL60 aux peptides histones acétylés. Il a une efficacité puissante contre les lignées cellulaires hébergeant différentes fusions MLL telles que les cellules MV4;11, RS4;11, MOLM13 et NOMO1 avec une IC50 de 15-192 nM. De manière cohérente, il ablate complètement le potentiel de formation de colonies des leucémies pilotées par la fusion MLL (MOLM13) mais pas les leucémies pilotées par l'activation de la tyrosine kinase (K562). I-BET151 présente également une efficacité puissante dans les essais de culture liquide et clonogéniques utilisant des progéniteurs murins primaires transformés avec MLL-ENL ou MLL-AF9. Le traitement avec ce composé induit significativement l'apoptose et un arrêt G0/G1 proéminent dans les lignées cellulaires de fusion MLL pilotées par des fusions MLL distinctes (MOLM13 et MV4;11 contenant MLL-AF9 et MLL-AF4, respectivement) mais pas les cellules K562, probablement en raison de l'inhibition de la transcription de BCL2, C-MYC et CDK6 en bloquant le recrutement des composants BRD3/4, PAFc et SEC au site de début de transcription (TSS).
|
| Kinase Assay |
Essai de déplacement de ligand par anisotropie de fluorescence (FP)
|
|
Tous les composants sont dissous dans un tampon de composition 50 mM HEPES pH 7.4, 150 mM NaCl et 0.5 mM CHAPS avec des concentrations finales de BRD 2/3/4 75 nM, ligand fluorescent 5 nM. 10 μL de ce mélange réactionnel sont ajoutés à l'aide d'un micro multidrop dans des puits contenant 100 nL de diverses concentrations de I-BET151 (GSK1210151A) ou de véhicule DMSO (1% final) dans une plaque de microtitration Greiner 384 puits noire à faible volume et équilibrés dans l'obscurité pendant 60 minutes à température ambiante. L'anisotropie de fluorescence est lue dans Envision (lex = 485 nm, lEM = 530 nm; Dichroïque = 505 nM).
|
|
| In vivo |
I-BET151 (GSK1210151A), administré à 30 mg/kg/jour, inhibe significativement la croissance tumorale des leucémies murines MLL-AF9 et humaines MLL-AF4 chez la souris, et procure un bénéfice de survie marqué.
|
Références |
Applications
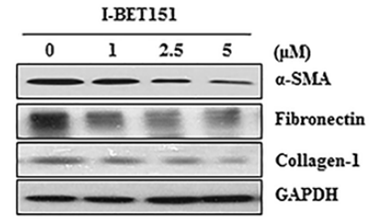
| Méthodes | Biomarqueurs | Images | PMID |
|---|---|---|---|
| Western blot | α-SMA / Fibronectin / Collagen-1 FoxM1 / AURKB / Survivin / cyclin B / PLK1 HP1α / HP1β / HP1γ |
|
27732564 |
Support technique
Tel: +1-832-582-8158 Ext:3
Si vous avez dautres questions, veuillez laisser un message.
