nur für Forschungszwecke
4-Methylumbelliferone (4-MU)
Kat.-Nr.S2256
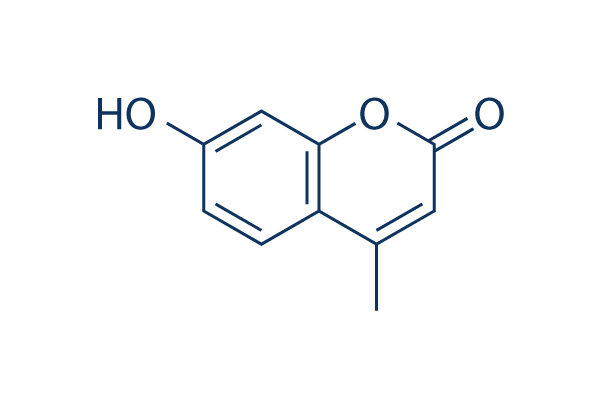
Chemische Struktur
Molekulargewicht: 176.17
Qualitätskontrolle
Zellkultur, Behandlung & Arbeitskonzentration
| Zelllinien | Assay-Typ | Konzentration | Inkubationszeit | Formulierung | Aktivitätsbeschreibung | PMID |
|---|---|---|---|---|---|---|
| COS1 cells | Function assay | Agonistic activity was determined in COS1 cells transfected with GAL 4-PPAR gamma receptor, EC50=0.021 μM | ||||
| CV-1 cells | Function assay | Maximal reporter activity against human Peroxisome proliferator activated receptor gamma Gal4 chimeric in transiently transfected CV-1 cells by functional assay, EC50=0.043 μM | ||||
| COS-1 cells | Function assay | In vitro transactivation of human peroxisome proliferator activated receptor gamma measured in PPAR-GAL4 chimeric COS-1 cells, EC50=0.02 μM | ||||
| CV-1 cells | Function assay | Activation of peroxisome proliferator activated receptor gamma measured by induction of 50% of maximum alkaline phosphatase activity, transfection assay in CV-1 cells, EC50=0.08913 μM | ||||
| CV-1 cells | Function assay | In vitro transcriptional activation of Peroxisome proliferator activated receptor gamma (PPAR) expressed in CV-1 cells, EC50=0.06 μM | ||||
| COS-1 cells | Function assay | In vitro agonist activity tested for transactivation in human PPAR gamma-Gal4 chimeric COS-1 cells, EC50=0.02 μM | ||||
| CV-1 cells | Function assay | In vitro evaluation against RXR-alpha/PPAR-gamma in CV-1 cells by cotransfection assay was determined, EC50=0.325 μM | ||||
| HepG2 cells | Function assay | Effective concentration against human PPARgamma expressed in HepG2 cells, EC50=0.039 μM | ||||
| Huh7 cells | Function assay | Functional activity at human PPAR gamma in Huh7 cells by transactivation assay, EC50=0.22 μM | ||||
| Huh7 cells | Function assay | Effect on PPAR gamma transactivation activity in Huh7 cells, EC50=0.22 μM | ||||
| U2OS cells | Function assay | Effect on PPARgamma transactivation activity in U2OS cells, EC50=0.03 μM | ||||
| CV1 cells | Function assay | Transactivation of PPARgamma in CV1 cells, EC50=0.076 μM | ||||
| human keratinocytes | Proliferation assay | Antiproliferative activity against human keratinocytes, EC50=8 μM | ||||
| HT29 cell | Proliferation assay | Antiproliferative activity against human HT29 cell line, EC50=45 μM | ||||
| NIH3T3 cells | Function assay | Activity at human PPAR gamma transfected in NIH3T3 cells by luciferase activity assay, EC50=0.32 μM | ||||
| CV1 cells | Function assay | Transactivation of human PPARgamma in CV1 cells by luciferase reporter gene assay, EC50=0.308 μM | ||||
| Huh7 cells | Function assay | Activity at human PPAR gamma in Huh7 cells by transactivation assay, EC50=0.22 μM | ||||
| HepG2 cells | Function assay | Agonist activity at human PPAR gamma in a HepG2 cells by PPAR-GAL4 transactivation assay, EC50=0.158 μM | ||||
| HepG2 cells | Function assay | Agonist activity at human PPAR gamma in HepG2 cells by PPAR-GAL4 transactivation assay, EC50=0.158μM | ||||
| CV1 cells | Function assay | Activity at human PPARgamma in CV1 cells, EC50=0.308 μM | ||||
| HEK293 cells | Function assay | Activity at human adipose tissue PPAR gamma expressed in HEK293 cells by PPAR-GAL4 transactivation assay, EC50=0.31 μM | ||||
| rat L6 cells | Function assay | Effect on fatty acid oxidation in rat L6 cells, EC50=5 μM | ||||
| HepG2 cells | Function assay | Agonist activity at human PPARgamma in HepG2 cells by PPAR-GAL4 transactivation assay, EC50=0.082 μM | ||||
| CV1 cells | Function assay | Agonist activity at human PPARalpha in CV1 cells by GAL4 transactivation assay after 24 hrs, EC50=3.46 μM | ||||
| CV1 cells | Function assay | Agonist activity at human PPARgamma in CV1 cells by GAL4 transactivation assay after 24 hrs, EC50=0.03 μM | ||||
| CV1 cells | Function assay | Agonist activity at human PPARgamma expressed in CV1 cells by receptor transactivation assay, EC50=0.308 μM | ||||
| CV1 cells | Function assay | Agonist activity at PPARgamma in CV1 cells by transactivation assay, EC50=0.308 μM | ||||
| HEK293 cells | Function assay | Agonist activity at PPARgamma receptor expressed in HEK293 cells by GAL4 transactivation assay, EC50=0.035 μM | ||||
| NIH3T3 cells | Function assay | Agonist activity at human PPARgamma expressed in NIH3T3 cells by GAL4 transactivation assay, EC50=0.32 μM | ||||
| HEK293 cells | Function assay | Agonist activity at PPARgamma expressed in HEK293 cells by GAL4 transactivation assay, EC50=0.19 μM | ||||
| HEK293 cells | Function assay | Agonist activity at PPARgamma expressed in HEK293 cells assessed as aP2 gene induction, EC50=0.12 μM | ||||
| HEK293 cells | Function assay | Agonist activity at PPARgamma in HEK293 cells by GAL4 transactivation assay, EC50=0.045 μM | ||||
| HepG2 cells | Function assay | Agonist activity at human PPARalpha ligand binding domain expressed in human HepG2 cells co-transfected with Gal4 by luciferase reporter gene assay, EC50=0.039 μM | ||||
| RAW264.7 cells | Function assay | Suppression of LPS/IFN-gamma-stimulated NO production in mouse RAW264.7 cells, EC50=17.5 μM | ||||
| BL21 cells | Function assay | Displacement of radio labeled 2(S)-(2-benzoyl-phenylamino)-3-{4-[1,1-ditritio-2-(5-methyl-2-phenyl-oxazol-4-yl)-ethoxy]-phenyl}-propionic acid from GST-fused human PPARgamma expressed in Escherichia coli BL21 cells by scintillation proximity assay, EC50=0.45 μM | ||||
| BHK21 cells | Function assay | Agonist activity at human PPARgamma expressed in BHK21 cells assessed as SEAP activity by luciferase reporter transactivation assay, EC50=0.45 μM | ||||
| HepG2/C3A cells | Function assay | Activation of human PPARgamma ligand binding domain-mediated transcriptional activity in human HepG2/C3A cells co-transfected with fused Gal4-LBD by cotransfection assay, EC50=0.0038 μM | ||||
| HepG2 cells | Function assay | Agonist activity at PPARgamma expressed in human HepG2 cells assessed as induction of receptor transactivation by reporter gene assay relative to control, EC50=0.05 μM | ||||
| COS1 cells | Function assay | Agonist activity at human recombinant PPARgamma expressed in COS1 cells co-expressing GAL4 assessed as transcriptional activity after 48 hrs by luciferase reporter gene assay, EC50=0.02 μM | ||||
| HepG2 cells | Function assay | Agonist activity at human PPARgamma expressed in HepG2 cells by GAL4 transactivation assay, EC50=0.223 μM | ||||
| CV1 cells | Function assay | Agonist activity at human PPARgamma ligand binding domain expressed in african green monkey CV1 cells co-transfected with fused Gal4-DBD by transactivation assay, EC50=0.033 μM | ||||
| CV1 cells | Function assay | Agonist activity at human PPARalpha ligand binding domain expressed in african green monkey CV1 cells co-transfected with fused Gal4-DBD by transactivation assay, EC50=3.46 μM | ||||
| U2OS cells | Function assay | Agonist activity at human PPARgamma in U2OS cells by transactivation assay, EC50=0.02 μM | ||||
| Hep G2 cells | Function assay | Agonist activity at human PPARgamma ligand binding domain expressed in human Hep G2 cells co-transfected with Gal4-DBD by luciferase reporter gene assay, EC50=0.04 μM | ||||
| HeLa cells | Function assay | Agonist activity at PPARgamma ligand binding domain expressed in human HeLa cells co-transfected with Gal4-DBD assessed as transcriptional activation by Gal4 response element-driven luciferase reporter gene assay, EC50=0.015 μM | ||||
| COS1 cells | Function assay | Agonist activity at human PPARgamma receptor expressed in african green monkey COS1 cells co-transfected with fused yeast Gal4-DBD by transactivation assay, EC50=0.02 μM | ||||
| U2OS cells | Function assay | Agonist activity at human PPARgamma expressed in U2OS cells by luciferase transactivation assay, EC50=0.05 μM | ||||
| HepG2 cells | Function assay | Agonist activity at GAL4-tagged human PPARgamma ligand binding domain expressed in human HepG2 cells assessed as receptor transactivation by luciferase reporter gene assay, EC50=0.039 μM | ||||
| HepG2 cells | Function assay | Agonist activity at human PPARgamma expressed in HepG2 cells by GAL4 transactivation assay, EC50=0.223 μM | ||||
| COS7 cells | Function assay | Agonist activity at human recombinant PPARgamma2 LBD expressed in african green monkey COS7 cells coexpressing GAL4 by luciferase reporter gene transactivation assay, EC50=0.1 μM | ||||
| COS7 cells | Function assay | Agonist activity at human recombinant PPARgamma1 LBD expressed in african green monkey COS7 cells coexpressing GAL4 by luciferase reporter gene transactivation assay, EC50=0.03 μM | ||||
| HEK cells | Function assay | Agonist activity at human GAL4-tagged PPARalpha chimeric receptor expressed in HEK cells by transactivation assay, EC50=10.58 μM | ||||
| HEK cells | Function assay | Agonist activity at human GAL4-tagged PPARgamma chimeric receptor expressed in HEK cells by transactivation assay, EC50=0.035 μM | ||||
| 3T3L1 cells | Function assay | 7 days | Induction of adipogenesis in mouse 3T3L1 cells assessed as increase in lipid accumulation after 7 days by Oil red O staining | |||
| CHO-K1 cells | Function assay | Partial agonist activity at human PPARgamma-LBD expressed in CHO-K1 cells co-transfected with GAL4 assessed as luciferase activity by transactivation assay, EC50=0.1 μM | ||||
| HEK293T cells | Function assay | 1 μM | 24 hrs | Partial agonist activity at human PPARgamma-LBD expressed in HEK293T cells assessed as induction of receptor transactivation at 1 uM after 24 hrs by luciferase reporter gene assay | ||
| CV1 cells | Function assay | Transactivation of Gal4-fused human PPARgamma DNA binding domain expressed in african green monkey CV1 cells by luciferase reporter gene assay, EC50=0.1 μM | ||||
| HEK293 cells | Function assay | 16 to 20 hrs | Transactivation of Gal4-fused human PPARgamma expressed in HEK293 cells after 16 to 20 hrs by luciferase reporter gene assay, EC50=0.043 μM | |||
| HepG2 cells | Function assay | 10 umol/L | 24 hrs | Increase in glucose consumption in human HepG2 cells at 10 umol/L after 24 hrs relative to control | ||
| 3T3L1 cell | Function assay | 1 uM | Induction of mouse 3T3L1 cell differentiation assessed as increase in accumulation of intracellular lipid droplet at 1 uM by Oil red O staining | |||
| CV1 cells | Function assay | 40 h | Agonist activity at human PPARgamma-LBD expressed in CV1 cells co-transfected with Gal4 after 40 hrs by luciferase based transactivation assay, EC50=0.1 μM | |||
| HepG2 cells | Function assay | Agonist activity at PPARgamma receptor expressed in human HepG2 cells by PPRE-luciferase reporter gene assay, EC50=0.01 μM | ||||
| HepG2 cells | Function assay | Transactivation of human PPARgamma expressed in human HepG2 cells co-transfected with PPRE3-TK-Luc by luciferase reporter gene assay, EC50=0.05 μM | ||||
| COS7 cells | Function assay | Modulation of human PPARgamma-LBD expressed in african green monkey COS7 cells co-transfected with Gal4 assessed as activation of transactivation activity by luciferase assay, EC50=0.043 μM | ||||
| COS-1 cells | Function assay | 24 hrs | Agonist activity at human PPARgamma ligand binding domain expressed in COS-1 cells after 24 hrs by luciferase reporter gene-based luminometric analysis, EC50=0.048 μM | |||
| COS1 cells | Function assay | 24 hrs | Transactivation of human full length PPARgamma expressed in COS1 cells co-transfected with RXRalpha after 24 hrs by luciferase reporter gene assay, EC50=0.12 μM | |||
| MG-63 cells | Function assay | Agonist activity at human PPARgamma expressed in MG-63 cells by reporter gene-based transactivation assay, EC50=0.011 μM | ||||
| COS1 cells | Function assay | Binding affinity to human wild type PPARgamma LBD expressed in COS1 cells co-expressing GAL4 by scintillation proximity assay | ||||
| MG-63 cells | Function assay | 24 hrs | Modulation of full-length human pSG5-fused PPARgamma expressed in MG-63 cells co-expressing pGV-P2-PPRE after 24 hrs by luciferase reporter gene based transactivation assay, EC50=0.011 μM | |||
| Ac2F cells | Function assay | 0.1 to 10 μM | 6 hrs | Agonist activity at PPARgamma in rat Ac2F cells assessed as luciferase activity at 0.1 to 10 uM after 6 hrs by reporter gene assay | ||
| 293H DA cells | Function assay | 16 h | Agonist activity at PPARgamma LBD in human 293H DA cells after 16 hrs by TR-FRET activation reporter assay, EC50=0.0024 μM | |||
| 3T3L1 cells | Function assay | 10 uM | Increase in adiponectin mRNA levels in TNFalpha-induced mouse 3T3L1 cells pretreated at 10 uM before TNF-alpha challenge relative to control | |||
| HepG2 cells | Function assay | 10 uM | 24 h | Reduction of glucose consumption in insulin-resistant human HepG2 cells at 10 uM after 24 hrs by glucose oxidase method in presence of 22.2 mM of glucose | ||
| HepG2 cells | Function assay | 10 uM | 24 h | Reduction of glucose consumption in insulin-resistant human HepG2 cells at 10 uM after 24 hrs by glucose oxidase method in presence of 0.1 uM of insulin | ||
| THP1 cells | Function assay | 100 uM | 3 hrs | Transactivation of PPARgamma in human THP1 cells assessed as decrease in LPS-induced MCP1 mRNA expression at 100 uM preincubated for 3 hrs prior to LPS challenge measured after 18 hrs by RT-PCR analysis relative to untreated control | ||
| THP1 cells | Function assay | 100 uM | 3 hrs | Transactivation of PPARgamma in human THP1 cells assessed as decrease in LPS-induced IL6 mRNA expression at 100 uM preincubated for 3 hrs prior to LPS challenge measured after 18 hrs by RT-PCR analysis relative to untreated control | ||
| HepG2 cells | Function assay | Transactivation of GAL4-fused PPARgamma ligand binding domain transfected in human HepG2 cells by luciferase reporter gene assay, EC50=0.1 μM | ||||
| HepG2 cells | Function assay | 20 h | Agonist activity at human GAL4-fused PPARgamma ligand binding domain expressed in HepG2 cells after 20 hrs by luciferase reporter gene transactivation assay, EC50=0.02 μM | |||
| BL21 DE3 cells | Function assay | Displacement of [3H]rosiglitazone from N-terminal His-tagged human PPARgamma ligand binding domain expressed in Escherichia coli BL21 DE3 cells by scintillation proximity assay, Ki=0.074 μM | ||||
| human L02 cells | Function assay | Agonist activity at PPARgamma-LBD expressed in human L02 cells co-expressing pGL3-SV40-GAL4 after 24 hrs by luciferase reporter gene based transactivation assay, EC50=0.9 μM | ||||
| HepG2 cells | Function assay | 20 h | Transactivation of GAL4-fused human PPARgamma ligand binding domain expressed in HepG2 cells after 20 hrs by luciferase reporter gene assay, EC50=0.039 μM | |||
| HEK293 cells | Function assay | 10 μM | Transactivation of PPARgamma (unknown origin) expressed in HEK293 cells co-transfected with AP2-PPRE at 10 uM by luciferase reporter gene assay | |||
| HEK293 cells | Function assay | Agonist at human PPARgamma LBD expressed in human HEK293 cells cotransfected with GAL4-Luc assessed as transcriptional activation by luciferase reporter gene assay, EC50=0.004 μM | ||||
| HEK293 cells | Function assay | 24 h | Transactivation of GAL4 DBD-fused human PPARgamma-LBD expressed in HEK293 cells after 24 hrs by luciferase reporter gene assay, EC50=0.1 μM | |||
| HEK293 cells | Function assay | 48 hrs | Agonist activity at human PPARgamma expressed in HEK293 cells cotransfected with PPREx4-TK-luc assessed as activation of luciferase activity measured after 48 hrs by transactivation assay, EC50=43.71 μM | |||
| HepG2 cells | Function assay | Agonist activity at GAL4-fused PPARgamma (unknown origin) expressed in human HepG2 cells by transactivation assay, EC50=0.01 μM | ||||
| HepG2 cells | Function assay | Agonist activity at GAL4-fused PPARgamma M463A mutant (unknown origin) expressed in human HepG2 cells by transactivation assay, EC50=0.002 μM | ||||
| L6 cells | Function assay | 24 hrs | Induction of glucose uptake in rat L6 cells pulsed with C14-deoxy glucose after 24 hrs in presence of insulin, EC50=4.49 μM | |||
| L6 cells | Function assay | 16 hrs | Increase in 2-[3H]-deoxyglucose uptake in rat L6 cells after 16 hrs by scintillation counting analysis, EC50=4.8 μM | |||
| DU145 cells | Cytotoxicity assay | 48 hrs | Cytotoxicity against human DU145 cells assessed as growth inhibition after 48 hrs by MTT assay, EC50=16 μM | |||
| PC3 cells | Cytotoxicity assay | 48 hrs | Cytotoxicity against human PC3 cells assessed as growth inhibition after 48 hrs by MTT assay, EC50=20.3 μM | |||
| MDA-MB-231 cells | Growth inhibition assay | 24 hrs | Growth inhibition of human MDA-MB-231 cells after 24 hrs by MTT assay, EC50=5.23 μM | |||
| HEK293 cells | Function assay | Agonist activity at human PPARgamma expressed in HEK293 cells by luciferase reporter gene assay, EC50=0.01 μM | ||||
| HepG2 cells | Function assay | 20 hrs | Transactivation of PPAR transfected in human HepG2 cells after 20 hrs by PPAR-luciferase assay, EC50=1.6 μM | |||
| HEK293 cells | Function assay | 24 hrs | Transactivation of PPARgamma (unknown origin) expressed in HEK293 cells incubated for 24 hrs by luciferase reporter gene assay | |||
| HepG2 cells | Function assay | Agonist activity at human GAL4-PPARgamma ligand binding domain expressed in human HepG2 cells by luciferase reporter gene assay, EC50=0.039 μM | ||||
| HEK293 cells | Function assay | 24 hrs | Agonist activity at mouse PPARgamma expressed in HEK293 cells co-expressing with Gal4 reporter vector after 24 hrs by dual-luciferase reporter assay, EC50=0.03 μM | |||
| HepG2 cells | Function assay | 20 hrs | Agonist activity at GAL4-DNA binding domain fused human PPARgamma ligand binding domain expressed in human HepG2 cells assessed as receptor transactivation incubated for 20 hrs by luciferase reporter gene assay, EC50=0.04 μM | |||
| HepaR cells | Function assay | 1 day | Agonist activity at PPARgamma in human HepaR cells assessed as increase in HMGCS2 gene expression at 1 uM incubated for 1 day by quantitative PCR method relative to untreated control | |||
| HepaR cells | Function assay | 1 uM | 1 day | Agonist activity at PPARgamma in human HepaR cells assessed as increase in ANGPTL4 gene expression at 1 uM incubated for 1 day by quantitative PCR method relative to untreated control | ||
| HepaR cells | Function assay | 1 uM | 1 day | Agonist activity at PPARgamma in human HepaR cells assessed as increase in FABP1 gene expression at 1 uM incubated for 1 day by quantitative PCR method relative to untreated control | ||
| HEK293 cells | Function assay | 10 μM | 24 hrs | Transactivation of PPAR-gamma (unknown origin) expressed in HEK293 cells at 10 uM after 24 hrs by luciferase reporter gene assay | ||
| C2C12 cells | Function assay | 30 μM | 2 h | Induction of 2-deoxy-[3H]-glucose uptake in mouse C2C12 cells at 30 uM after 2 hrs by liquid scintillation counting analysis | ||
| MCF7 cells | Function assay | 16 h | Agonist activity at human PPARgamma transfected in human MCF7 cells after 16 hrs by luciferase reporter gene assay, EC50=0.087 μM | |||
| HEK293 cells | Function assay | 10 μM | 24 h | Agonist activity at PPARgamma (unknown origin) expressed in HEK293 cells assessed as receptor transactivation at 10 uM incubated for 24 hrs by luciferase reporter gene assay | ||
| Klicken Sie hier, um weitere experimentelle Daten zu Zelllinien anzuzeigen | ||||||
Chemische Informationen, Lagerung & Stabilität
| Molekulargewicht | 176.17 | Formel | C10H8O3 |
Lagerung (Ab dem Eingangsdatum) | |
|---|---|---|---|---|---|
| CAS-Nr. | 90-33-5 | SDF herunterladen | Lagerung von Stammlösungen |
|
|
| Synonyme | N/A | Smiles | CC1=CC(=O)OC2=C1C=CC(=C2)O | ||
Löslichkeit
|
In vitro |
DMSO
: 35 mg/mL
(198.67 mM)
Ethanol : 35 mg/mL Water : Insoluble |
Molaritätsrechner
|
In vivo |
|||||
In-vivo-Formulierungsrechner (Klare Lösung)
Schritt 1: Geben Sie die untenstehenden Informationen ein (Empfohlen: Ein zusätzliches Tier zur Berücksichtigung von Verlusten während des Experiments)
Schritt 2: Geben Sie die In-vivo-Formulierung ein (Dies ist nur der Rechner, keine Formulierung. Bitte kontaktieren Sie uns zuerst, wenn es im Abschnitt "Löslichkeit" keine In-vivo-Formulierung gibt.)
Berechnungsergebnisse:
Arbeitskonzentration: mg/ml;
Methode zur Herstellung der DMSO-Stammlösung: mg Wirkstoff vorgelöst in μL DMSO ( Konzentration der Stammlösung mg/mL, Bitte kontaktieren Sie uns zuerst, wenn die Konzentration die DMSO-Löslichkeit der Wirkstoffcharge überschreitet. )
Methode zur Herstellung der In-vivo-Formulierung: Nehmen Sie μL DMSO Stammlösung, dann hinzufügenμL PEG300, mischen und klären, dann hinzufügenμL Tween 80, mischen und klären, dann hinzufügen μL ddH2O, mischen und klären.
Methode zur Herstellung der In-vivo-Formulierung: Nehmen Sie μL DMSO Stammlösung, dann hinzufügen μL Maisöl, mischen und klären.
Hinweis: 1. Bitte stellen Sie sicher, dass die Flüssigkeit klar ist, bevor Sie das nächste Lösungsmittel hinzufügen.
2. Achten Sie darauf, das/die Lösungsmittel der Reihe nach hinzuzufügen. Sie müssen sicherstellen, dass die bei der vorherigen Zugabe erhaltene Lösung eine klare Lösung ist, bevor Sie mit der Zugabe des nächsten Lösungsmittels fortfahren. Physikalische Methoden wie Vortex, Ultraschall oder ein heißes Wasserbad können zur Unterstützung des Lösens verwendet werden.
Wirkmechanismus
Klinische Studieninformationen
(Daten von https://clinicaltrials.gov, aktualisiert am 2024-05-22)
| NCT-Nummer | Rekrutierung | Erkrankungen | Sponsor/Kooperationspartner | Startdatum | Phasen |
|---|---|---|---|---|---|
| NCT05295680 | Recruiting | Primary Sclerosing Cholangitis |
Aparna Goel|Stanford University |
May 10 2023 | Phase 2 |
Technischer Support
Tel: +1-832-582-8158 Ext:3
Wenn Sie weitere Fragen haben, hinterlassen Sie bitte eine Nachricht.
